Most difficult is the labeling by other people that models are empty-headed bodies. Boyd Holbrook
Prerequisite
Data with headshape points (3 fiducials, and ~100 head points, optionally 4 HPI coils) should have been digitized before MEG data acquisition.
Individual subject MRI should have been acquired.
Goal of this tutorial
Merge (i.e. realign) the sensor locations of the MEG recordings with the MRI of the individual subject, create a head model and a grid (aka leadfield matrix) for source analysis.
Step-by-step guide
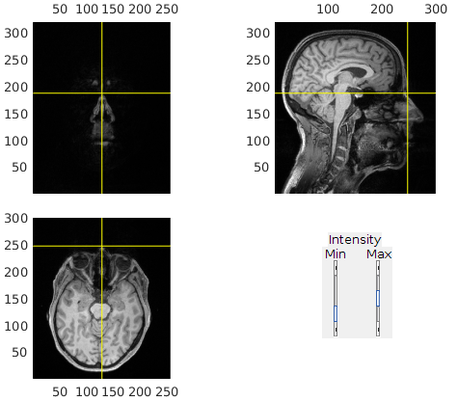
1. Load the MRI and "reslice" it
In this step, we simply flip the MRI so it is upright. Skipping this step will result in mixed up slices and/or upside down image.
mrif = '/path/and/filename/to/T1_mri.mgz'; % the file format could be one of many supported by fieldtrip. Native DICOM format works but is a bit slow to load. % read in the mri mri=ft_read_mri(mrif); % flip image so it looks upright cfg = []; cfg.method = 'flip'; mri_aligned = ft_volumereslice(cfg,mri);
using freesurfer's orig file
Note that if you have used freesurfer previously to segment your MRI, you can load the original mri stored in your freesurfer subjects' directory under SubjectName/orig/001.mgz, which will load faster than the DICOMs
2. Align the MRI with the fiducial points
This step is just a rough alignment of the MRI fiducial points. The next step will use head shape points to refine this alignment. If no head shape points have been digitized, it is important to point precisely to the fiducial points here.
% realign with fiducials cfg = []; cfg.method = 'interactive'; cfg.coordsys = 'neuromag'; mri_aligned = ft_volumerealign(cfg, mri_aligned);
Nasion (press n)
Left preauricular (LPA, press l)
Right preauricular (RPA, press r)
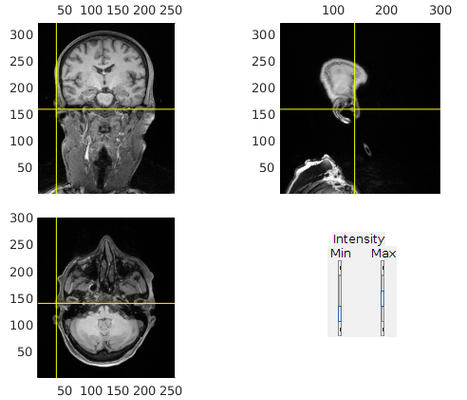
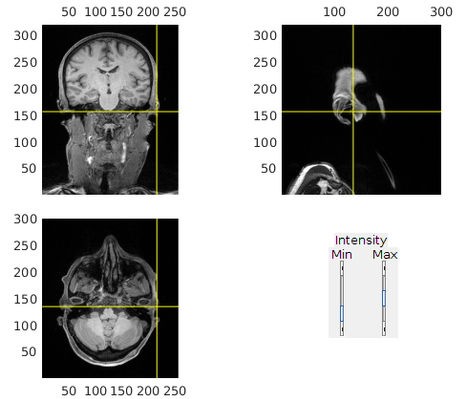
3. Refine alignment with head shape points
% headshape should be stored along with the MEG data headshapef = '/path/to/MEGdata.fif'; cfg = []; cfg.method = 'headshape'; headshape = ft_read_headshape(headshapef); headshape = ft_convert_units(headshape, 'mm'); cfg.headshape.headshape = headshape; cfg.coordsys = 'neuromag'; cfg.headshape.interactive = 'no'; mri_aligned = ft_volumerealign(cfg, mri_aligned);
This step can also be done interactively to further refine the alignment (set cfg.headshape.interactive = 'yes').
4. Create a head model (3D surfaces for brain, skull and scalp)
Segment and extract brain, skull and scalp surfaces.
cfg = [];
cfg.output = {'brain','skull','scalp'};
mri_seg = ft_volumesegment(cfg, mri_aligned);
cfg = [];
cfg.method = 'singleshell';
cfg.tissue = {'brain','skull','scalp'};
headmodel = ft_prepare_headmodel(cfg,mri_seg);
Surfaces
Note that only the scalp surface is necessary to create the head model for source localization. It looks much nicer on the upcoming figures with 3 layers, though.
5. Load and align a template grid to the head model
cfg = []; cfg.grid.warpmni = 'yes'; cfg.grid.template = template_grid; cfg.grid.nonlinear = 'yes'; cfg.grid.unit = 'mm'; cfg.mri = mri_aligned; grid = ft_prepare_sourcemodel(cfg); % save the results save(fullfile(sujdir,'source_head_model.mat'), 'mri', 'mri_aligned','mri_seg','headmodel','grid','headshape');
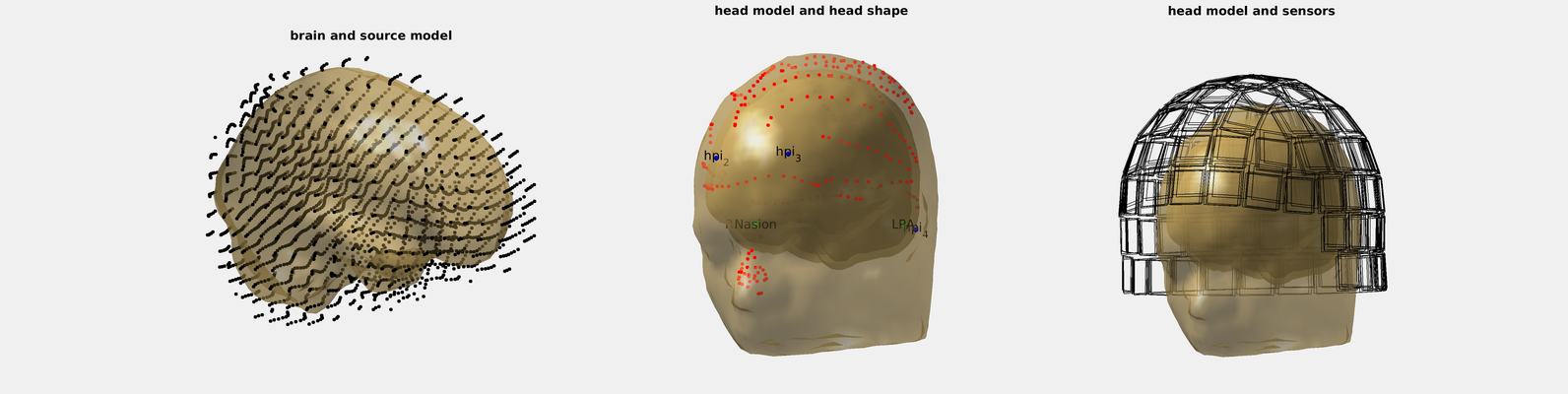
6. Plot the results and check
% make a figure with enough space for 3 panels horizontally
p = get(0,'defaultfigureposition'); p(3) = p(3) * 3;
figure('name',suj,'numbertitle','off','position',p);
% first panel with brain and aligned grid
subplot(131);
hm = headmodel;hm.bnd = hm.bnd(1);% use only the first layer (brain)
ft_plot_vol(hm,'vertexcolor','none','facecolor','skin','edgecolor','none','facealpha',.5);
ft_plot_mesh(grid.pos(grid.inside,:));%
lighting gouraud
material shiny
camlight
view(90,0)
rotate3d on
drawnow
title('brain and source model')
% second panel with head shape points, HPI coils, scalp, skull, and brain.
subplot(132);
ft_plot_vol(headmodel,'vertexcolor','none','facecolor','skin','edgecolor','none','facealpha',.5);
ft_plot_headshape(headshape);
hpi = find(cellfun(@(x)~isempty(x),regexp(headshape.label,'hpi_.*')));
hold on
scatter3(headshape.pos(hpi,1),headshape.pos(hpi,2),headshape.pos(hpi,3),200,'b','.');
text(headshape.pos(hpi,1),headshape.pos(hpi,2),headshape.pos(hpi,3), headshape.label(hpi), 'HorizontalAlignment', 'center', 'VerticalAlignment', 'middle', 'Interpreter', 'tex');
lighting gouraud
material shiny
camlight
view(-150,0)
rotate3d on
drawnow
title('head model and head shape')
% third panel with head and MEG sensors
subplot(133);cla
ft_plot_vol(headmodel,'vertexcolor','none','facecolor','skin','edgecolor','none','facealpha',.5);
lighting gouraud
material shiny
camlight
view(-180,0)
drawnow
% Below is the list of all runs by this subject
allruns = {'run01_tsss.fif','run02_tsss.fif','run03_tsss.fif'};
for i_run = 1:numel(allruns)
h = ft_read_header(allruns{i_run});
h.grad = ft_convert_units(h.grad, 'mm');
ft_plot_sens(h.grad);
end
rotate3d on
drawnow
title('head model and sensors')
drawnow
% save as image and as .fig
export_fig(hfig,fullfile(rootdir,'figures','headmodel',['headmodel_' suj '.png']),'-nocrop','-r300'); % https://github.com/altmany/export_fig
saveas(hfig,fullfile(rootdir,'figures','headmodel',['headmodel_' suj '.fig']))
At this point we recommend to check (use the mouse to rotate the heads):
- The grid spans the whole brain and about a cm beyond
- The head shape points are aligned to the scalp surface
- Nasion, LPA and RPA are where they are supposed to be (although the head shape realignment moved them from where we clicked earlier)
- HPI coils are in place
- Sensors are where they are supposed to be (note that if you transformed the coordinate frame with Maxfilter to one recording run, the sensors should superimpose exactly, otherwise not)
Related articles
Add Comment