This is a collaborative space. In order to contribute, send an email to maximilien.chaumon@icm-institute.org
On any page, type the letter L on your keyboard to add a "Label" to the page, which will make search easier.
Using Maxfilter
Prerequisite
Maxfilter is applied to raw MEG data just after recording on .fif files. It is usually the very first preprocessing step applied (except perhaps merging several data modalities).
Goal of this tutorial
This tutorial teaches how to use the Maxfilter command line tool to remove artifacts on MEG signals coming from outside of the sphere of all MEG channels. This tool was build by Elekta, the vendor of our MEG.
It is also at this step that bad channels are marked and discarded (interpolated).
Step-by-step guide
- Choose a reference head position
- Run Maxfilter on the raw data
- Check correction with Muse
- Iterate to fine tune correction parameters
sss_config folder
Before running Maxfilter, ensure that there is a sss_config folder next to the recordings. This folder contains important parameters for Maxfilter to run properly. If missing, it will be silently ignored.
Choose head position
Even though not mandatory, this step is important because it facilitates merging different runs in subsequent steps, especially if you compute evoked fields over several runs. Typically, one (the first) run or the mean position of the session will be used as reference. This head position is passed to maxfilter via the -trans option.
If you choose to use a mean head position to realign during the tsss processing, it can be computed as follows:
dataHandler -r -avg run01.fif run02.fif run03.fif run04.fif -hp mean -time 0 -.1 meanhp.fif
This will average head positions in run 01,02,03,04 and save it to meanhp.fif, which can be used as input to the -trans option below.
Run Maxfilter
Here is a generic example of a Maxfilter command at CENIR:
maxfilter_cenir -f run01.fif -st 20000 -corr 0.98 -bad 0221 0332 -trans meanhp.fif -o run01_tsss.fif
Brief description of the options (the user is referred to the pdf manual for more information):
-f: the input file-o: the output file-tsss: perform temporal source space separation (the standard at CENIR). The option is followed by a buffer length (in seconds), which should always be larger than your individual recordings.-corr: amount of correlation between data and noise that should be rejected (see manual for more explanation). Lowering the value below the default 0.98 may sometimes help getting rid of artifacts but can also lead to removing interesting signal from the data. Careful inspection is recommended (see below).-bad: which channels should be considered bad, i.e. removed prior to correction and later reinterpolated. The -autobad command is not recommended because it leaves no trace of which channels are considered bad. Rather use xscan (see below).-trans: transforms the MEG data into the sensor array defined by the channel info in a specified file (heremean.fif).
use maxfilter_cenir
Be sure to use the maxfilter_cenir command and NOT maxfilter. This will ensure that correct tuning parameters found in the sss_config directory next to your data are applied during the computation. The full script below spells out the full command for each file it runs on.
In case of large artifacts, be sure to check this
On rare occasions, we experienced some trouble with tsss processing: when large dc-offset is present in the signal at the beginning of the file a bug can occur, leaving the resulting file with strong artifacts at the beginning of the file and a lot of noise above 80Hz on a number of channels.
dataHandler -rbr 1 1000 -new run02_dc_offset run02.fiff
maxfilter_cenir -gui -st 20000 -trans meanhp.fif -f run02_dc_offset.fif
Automated bad channel detection
Maxfilter has an -autobad option to detect noisy or defect channels. We do not recommend using this option because it does not log which channels were considered bad and thus prevents tracability. We rather recommend using Elekta's xscan command which allows listing bad channels. The usage of this command is very similar to that of maxfilter. Run xscan -help for more information. The script below uses xscan to list bad channels.
Full script
The script below takes any number of .fif files as input and generates up to three outputs per file.
For instance, for a file called file1.fif, it creates:
1/ a maxfiltered version file1_tsss.fif (using maxfilter -st)
2/ a list of bad channels file1.bad (using xscan). If a file file1.bad already exists, this step is skipped and the content of file1.bad will be used for step 3.
3/ a maxfiltered version excluding these bad channels file1_tsss_nobad.fif (using maxfilter -bad )
If the -trans fileX.fif option is passed (often recommended), the head position in each file will be
set to that in fileX.fif
Copy and paste the code below in a text file (following instructions here) to create a script.
If we call the script above "maxfilter.script", we run it as follows:
./maxfilter.script /path/to/mydata/file1.fif /path/to/mydata/file2.fif -trans /path/to/mydata/file1.fif
This will process two files file1.fif and file2.fif in the folder called /path/to/mydata and set the position of the head to that in file1.fif
Maxfilter user's manual
Further information on Maxfilter's usage can be found here
Inspect with muse
After Maxfilter has finished, it is important to check signal quality visually. We use Muse to do this.
muse run01_tsss.fif
Important aspects of the data to pay attention to:
- "ticks" on all sensors → one electronic artifact happened on a sensor that hasn't been treated as bad. Usually this is the sensor on which the amplitude of the "ticks" are maximal.
- Whether or not large eye artifacts have been removed. The eyes are at the limit of what Maxfilter considers the head sphere. It may consider that blinks originate from outside the sphere and discard them. Whether you want that or not is your call.
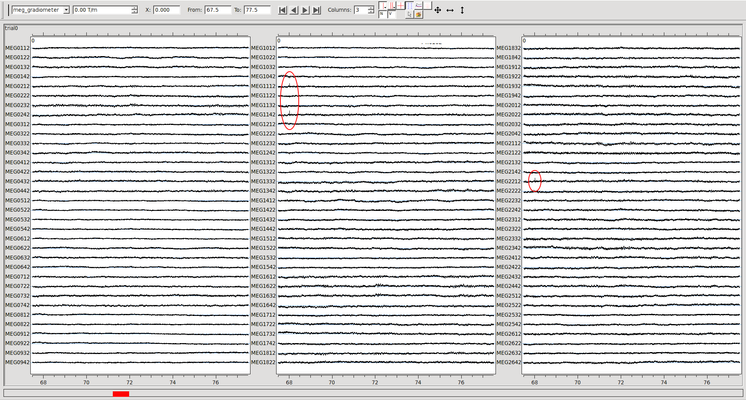
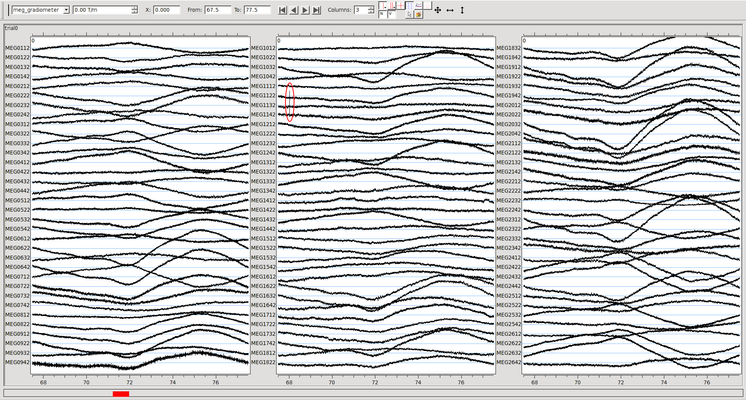
Below is an example of a file that has been processed without removing a bad channel. The very brief artifact highlighted in red occurred on only one sensor. Maxfiltering has spread it to several others. (Click on the image for a larger version).
Below is the same data before any maxfiltering. The artifact occurs on only one channel (1142, zooming with Ctrl+mouse wheel would reveal that only one channel is involved). The artifact that is present above on channels 2232 and 1112 is not visible here. Not discarding it during maxfilter above "spread" this artifact to other channels.
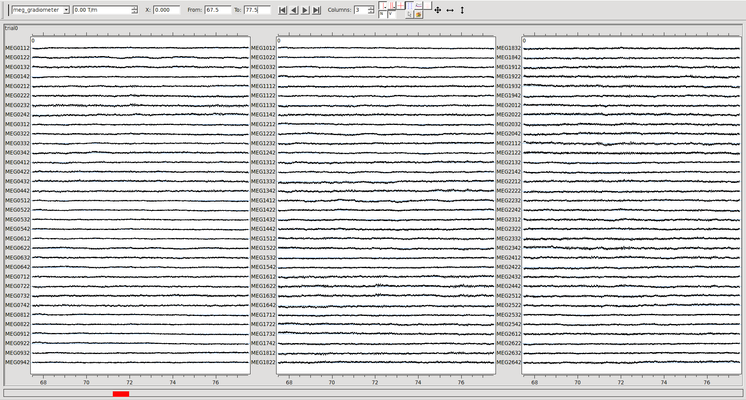
Below is the same data where channel 1142 has been marked as bad (with the command -bad 10142). Note that this bad channel was detected with the automatic xscan command included in the script above.
Related articles