This is a collaborative space. In order to contribute, send an email to maximilien.chaumon@icm-institute.org
On any page, type the letter L on your keyboard to add a "Label" to the page, which will make search easier.
FreeSurfer segmentation
Former user (Deleted)
Laurent HUGUEVILLE
Maximilien CHAUMON
Prerequisite
Anatomical MRI should have been acquired. The MRI sequence is described in the document on the right.
Goal of this tutorial
We will explain how to run a brain segmentation using FreeSurfer for source and head model design to analyse MEG data.
FreeSurfer finds interfaces between various brain structures (white & gray matter, gray and CSF...) and creates 3D models of these interfaces for later import in an MEG analysis software.
FreeSurfer Analysis Pipeline Overview is a good place to start tackling these steps (also useful info in the FAQ). Below is a minimalist setup guideline which should work at the CENIR for most subjects.
Step-by-step guide
Setup and run FreeSurfer
We set environment variables as follows.
$ export FREESURFER_HOME=/network/lustre/iss01/cenir/software/meeg/freesurfer/ $ export SUBJECTS_DIR=/network/lustre/iss01/cenir/analyse/meeg/YOUR_PROJECT/subjects $ export SUBJECT=S01
We refer to these variables throughout this tutorial.
The segmentation program is called recon-all.
$ recon-all -i one_dicom_file_for_that_subject.MR.dic -all -qcache
This will run overnight. Come back tomorrow.
Results
Once computations are done, we need to check that the segmentation finished without error.
In the results folder ($SUBJECTS_DIR/$SUBJECT), there is a scripts directory and a file called recon-all-status.log.
This file should end with something similar to this:
#@# Qdec Cache rh w-g.pct.mgh fwhm20 fsaverage Sat Mar 25 02:33:49 CET 2017 #@# Qdec Cache rh w-g.pct.mgh fwhm25 fsaverage Sat Mar 25 02:33:54 CET 2017 #@# BA Labels lh Sat Mar 25 02:34:00 CET 2017 #@# BA Labels rh Sat Mar 25 02:37:42 CET 2017 #@# Ex-vivo Entorhinal Cortex Label lh Sat Mar 25 02:41:24 CET 2017 #@# Ex-vivo Entorhinal Cortex Label rh Sat Mar 25 02:41:36 CET 2017 recon-all -s S01 finished without error at Sat Mar 25 02:41:49 CET 2017
Important is the mention: finished without error .
Checking the results
Once the segmentation is done, we can view it with various tools:
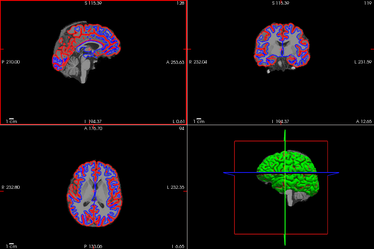
Freeview
The simplest method is using Freeview. (you may need to install this library or a similar one)
$ source /network/lustre/iss01/cenir/software/irm/bin/source_sepproject.bash $ fsViewPatient.py $SUBJECT
or
$ fsViewPatient.py $SUBJECT -a
These are equivalent to the command:
freeview -v ${SUBJECTS_DIR}/${subject}/mri/orig.mgz \
${SUBJECTS_DIR}/${subject}/mri/wm.mgz \
${SUBJECTS_DIR}/${subject}/mri/brainmask.mgz \
${SUBJECTS_DIR}/${subject}/mri/aparc+aseg.mgz \ # this line is added with the -a option above
-f ${SUBJECTS_DIR}/${subject}/surf/lh.pial:edgecolor=red \
-${SUBJECTS_DIR}/${subject}/surf/lh.white:edgecolor=blue \
-${SUBJECTS_DIR}/${subject}/surf/rh.pial:edgecolor=red \
-${SUBJECTS_DIR}/${subject}/surf/rh.white:edgecolor=blue
Which loads:
- volumes (-v) orig.mgz, wm.mgz and brainmask.mgz
- and surfaces (-f) pial surface in red, white-gray matter interface in blue
- The aparc+aseg volume containing automated classification (labelling) of subcortical structuresChecking consists principally in verifying that surfaces are well aligned (non overlapping), and stick to the expected contrasts in the MRI.
If a problem appears, first check this page: TroubleshootingData.
Then there are usually two options:
Adjust parameters and restart the pipeline (entirely or in part). Options
-wsthreshand-no-wsgcaatlasare worth your attention (SkullStripFix_freeview, also check the complete list of options:recon-all -help).Editing brainmask by hand.
In any case, detailed information can be found at TroubleshootingData.
Tksurfer and Tkmedit
Another method uses tksurfer and tkmedit. Here are some of the possible checks, which should be adjusted to meet the specific needs of the user.
# checking surfaces $ tksurfer $SUBJECT lh inflated $ tksurfer $SUBJECT rh inflated # check the talairach transform tkregister2 --mgz --s $SUBJECT --fstal --surf orig # check the skull strip tkmedit $SUBJECT brainmask.mgz -aux T1.mgz -surfs # check the white and pial surfaces tkmedit $SUBJECT brainmask.mgz -aux wm.mgz -surfs # check the segmentations tkmedit $SUBJECT brainmask.mgz -surfs -aseg
For more detailed documentation (instructions, advice, examples): RecommendedReconstruction.
For details on manual editing of segmented surfaces: TkMeditSegmentations.