This is a collaborative space. In order to contribute, send an email to maximilien.chaumon@icm-institute.org
On any page, type the letter L on your keyboard to add a "Label" to the page, which will make search easier.
Time-locked analysis - Sources
In this tutorial you will find a description of all the steps necessary to obtain the sources localisation of your evoked potentials / fields using a weighted Minimum Norm Approach (wMNE) in BrainStorm.
Prerequisites
- Preprocessing is complete
- T1 MRI data is preprocessed (segmented) or you chose a template
- Download the latest version of BrainStorm - Install it in your software folder - Take a look at the BrainStorm tutorials!
Input data
Evoked potentials (EEG)
Brainstorm can import the data coming from our BrainAmps system. If a 3D digitalisation of the EEG electrodes coordinates was performed you should insert this information in the file using multiconv (see the appropriate tutorial). It is also possible to use an EEG template (you can do that after the EEG importation in BrainStorm).
Evoked fields (MEG)
BrainStorm can import the fif files coming from our MEG system and those pre processed with our in-house tools. If you acquired simultaneous MEG+EEG data, BrainStorm will import both of them.
Noise covariance data
You will need to compute the "noise covariance matrix" used to attenuate what you consider as "noise" in your signal. For this you need to provide the corresponding data. It can be only pure sensor noise (without subject) or a specific part of the recording with the subject (a "baseline"). For instance, it can be a pre stimulus period, a separate resting-state recording.
Same preprocessing!
Note that, both the signal used to compute the evoked fields / potentials and the noise covariance matrix should be preprocessed in the same way (tsss, artefact removal or rejection, filtering)
MEG
We have a direct access to the sensor noise level since it is quite easy to perform an empty room recordings. Depending on your experimental setup and your scientific questions you can use either an empty room recording (useful when analysing resting state data), a resting state recording or a pre stimulus window when you are analysing a task. Be careful, all the activities occurring in the chosen signal will be attenuated during the localisation process.
Empty room and resting state recordings
For each subject do a 5 minutes empty room recording during the subject preparation and a 5 minutes eye open resting state recording at the beginning of the session
EEG
We have not an easy access to the sensor noise level (no easy empty room recordings!). Depending on your experimental setup and your scientific questions you can use either an identity matrix (useful when analysing resting state data), a resting state recording or a pre stimulus window when you are analysing a task. Be careful, all the activities occurring in the chosen signal will be attenuated during the localisation process.
Resting state recordings
For each subject do a 5 minutes eye open resting state recording at the beginning of the session
For more detailed information read this tutorial: Noise and data covariance matrices
MRI
If you have the individual anatomy of each of your subjects (T1 MRI), you should process it with FreeSurfer. The resulting folder will be imported for each subject into BrainStorm.
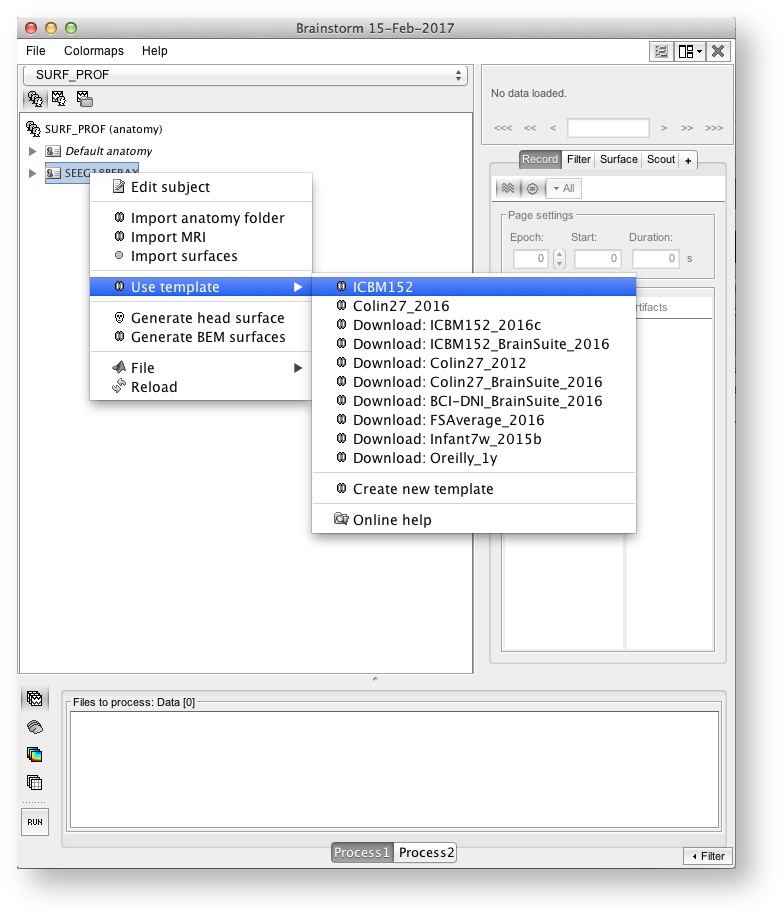
If you don't have the individual anatomy, then you have to choose a template in BrainStorm (the default one, ICBM 152, will do the job).
Needed software
wMNE localisation with BrainsStorm
Database folder
Create a folder DB_BST in your results folder and then use it to store your protocole when following this tutorial: Create a new protocol
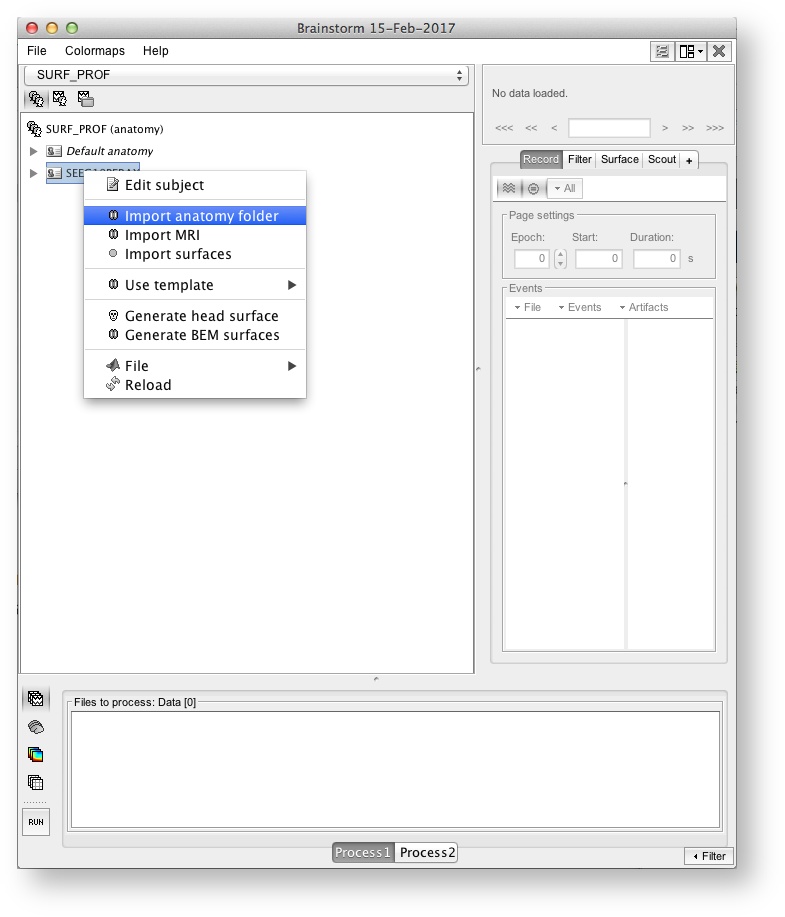
Create your subjects and import the anatomy
Import the individual MRI data (processed in freesurfer - Use "Import anatomy folder" menu item)
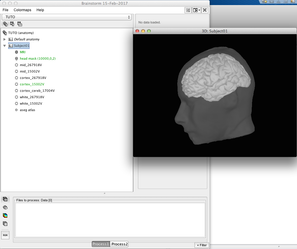
The end product is a complete description of the anatomy of the subject (left panel) + a figure with a 3D view of the subject's skin + cortex
If you have no MRI data: Choose and use a template (ICBM 152, the old colin27, will do the job)
Fiducial points
Each time you import the anatomy of a subject (or choose a template) you have to specify the "Fiducial points" used to register MRI data to MEG/EEG data, be careful to place them correctly on the MRI! This will impact the accuracy of the registration and thus it will impact the accuracy of your whole localisation process!
Import your evoked field
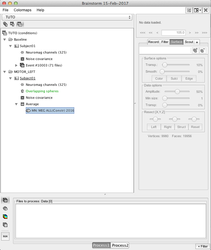
First step: Create your conditions by using the "View by conditions panel"
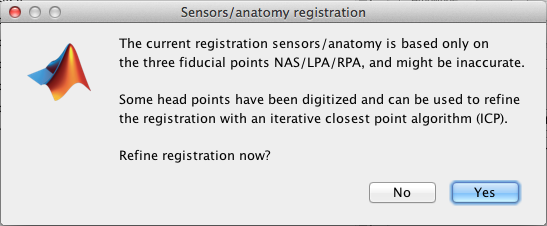
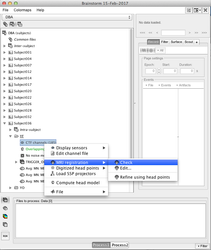
Use the menu item "Import MEG/EEG " to import your fif evoked fields (for each subject, each condition). Each time, answer "yes", when BrainStorm proposes to Refine the registration - This will insure the best MRI to MEG registration possible (see: Channel file / MEG-MRI coregistration)
As described in the BrainStorm tutorial, check the MRI-MEG registration!
Import your evoked potentials
Use the menu item "Import MEG/EEG" to import your evoked potentials (for each subject, each condition). If you added the electrodes localisation during the preprocessing, the EEG to MRI registration should be done automatically (check it!), if you don't have the electrodes coordinates, then you have to choose an EEG template as described in this tutorial.
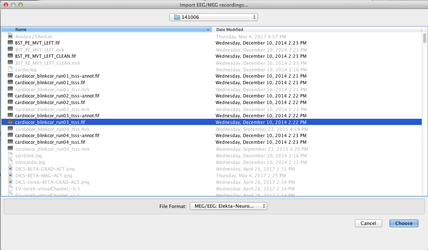
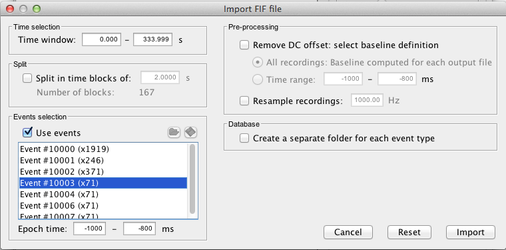
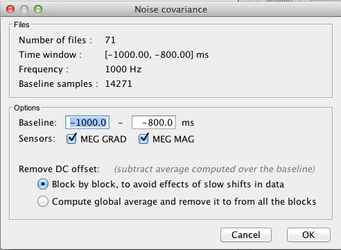
Import the data for the noise covariance matrix
Use the menu item "Import MEG/EEG" to import the data to be used for noise data covariance matrix. Here is an exemple where we imported 71 trials of 200 ms 1 second before a given marker.
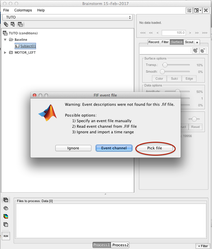
Select the pre processed signals (NOT the averaged one!):
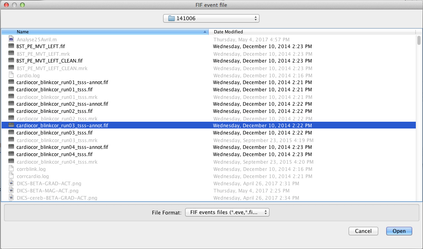
Select the chosen time window (here a window around a marker piked in the -annot.fif file):
→ →
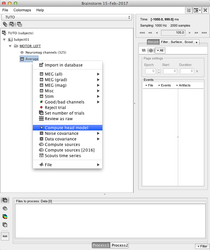
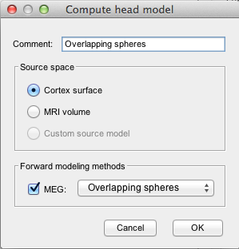
Compute the head model
Compute the head model (i.e. the forward model, the forward operator):
→
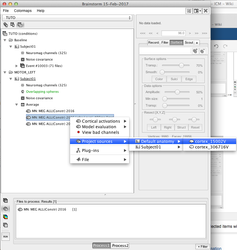
Processing multiple conditions and subjects
You can select several conditions or several subjects, do a right-click, and launch the head model computation for all the selected items with one-click
More information in: Head modeling
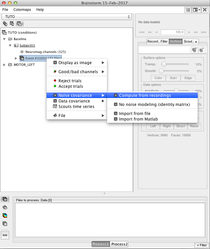
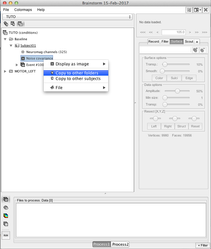
Compute the noise covariance matrix
When all conditions for one subject are imported, you can compute the noise covariance matrix and copy it into each condition:
→ → →
More information in: Noise and data covariance matrices
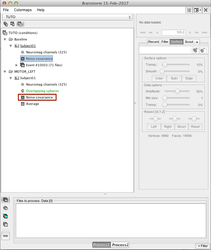
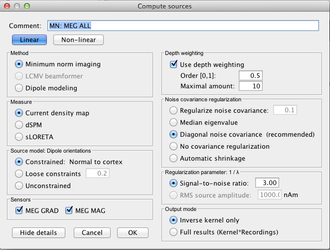
Compute the sources
One you have the head model and the noise covariance matrix, you compute the sources:
→ →
Processing multiple conditions and subjects
You can select several conditions or several subjects, do a right-click, and launch the sources computation for all the selected items with one-click
For more information concerning source localisation and visualisation of the results read the following tutorial: Source estimation
Projection to a template and Group analysis
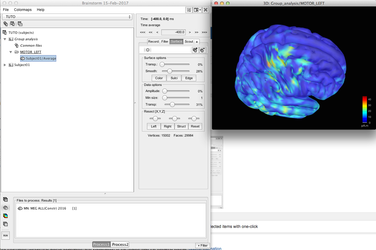
When the sources have been computed at the subject level, it is possible to project the results to a common space, i.e. a template, generally the default anatomy defined for the protocole:
→
Using these projected sources you can analyse the results at the group level using the process window:
→
Use filter
You can use "filter" in the process window to select files to be analysed at one point (above we selected an average from -400ms to -200ms). This will ease the files selection in the group results.
Advanced topics
Do I use an orientation constraint?
Good question, in a perfect world, with a perfect description of the subject anatomy and, main point, a perfect MEG/EEG to MRI registration, the answer is yes.
However, all the prerequisites may not be ok in your case. Here is a set of rules to make a decision, knowing that you can easily do both a constrained and an unconstrained localisation in BrainStorm.
Constrained vs Unconstrained
Subject individual MRI + good registration → Yes
Subject individual MRI + bad registration → No
MRI Template → No
BUT
Constrained vs Unconstrained
Without constraint → you will get three components per sources -> If you use the norm (x2+y2+z2) time-Frequency / causality analysis are impossible (instead you may do a pca, but you will a part of the information)
Read the full discussion here: Sign of constrained maps + Unconstrained orientations
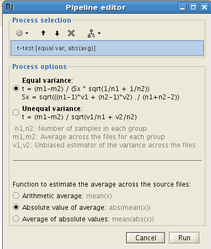
Doubt about current density units: pA.m or pA/m2?
wMNE results are expressed as paA.m and not pA/m^2 - why ?
From John Mosher:
Short answer:
Brainstorm currently presents each grid point as a current dipole, in units of A-m. We may infer surface density by dividing the dipole strength by the implied area around each dipole, and we may infer volume density by additionally dividing out the implied cortical thickness.
So if we run the Brainstorm Median Nerve Sample as a minimum norm, for 15,000 vertices, we see peaks of about 500 pA-m around 35ms post-stimulus. If we reducepatch the cortex to 1,500 vertices, we see peaks of 5,000 pA-m. Under the Scouts, if we select all clusters, Brainstorm reports in both cases a cortical surface area of about 2000 cm^2. Therefore each dipole in the 1500 dipole model represents ten times the area of the 15,000 dipole model, hence each dipolar moment is 10x greater.
As a rough approximation, for min norm models employing 15,000 vertices in an adult model, then the rough conversion would be 500 pA-m => 50 nA/mm surface density (see below calculations).
Long answer: Search for Doubt about current density units: pA.m or pA/m2? in the brainstorm forum
DBA analysis