Cherish your human connections. (Barbara Bush)
The Human Connectome Project is a large scale open data collection. It has an MEG component.
Resources:
Human Connectome Project: http://www.humanconnectomeproject.org/
MEG pipeline: https://www.humanconnectome.org/software/hcp-meg-pipelines
Data download : Only needed if you cannot find the data locally. https://db.humanconnectome.org (Does not work with Firefox (April 2018)) We recommend getting in touch with Maximilien CHAUMON instead.
FAQ: https://wiki.humanconnectome.org/display/PublicData/HCP+Users+FAQ
Prerequisite
We hold a local copy of a good portion of the MEG data of the HCP. This data is located on the network at /this/address/ in original zip (archive) format. If the data you are looking for is not at this address, please contact Maximilien CHAUMON.
Goal of this tutorial
Create simple code to extract parts of the HCP data to a different folder.
Step-by-step guide
All you need to do to extract the data is to:
- Locate the zip files, open one of them and look for the data you need
- List the zip files (e.g. for all subjects)
- Unzip what you need locally.
Locate the files you need. These are named, according to the HCP nomenclature. For instance, the resting state data is named like this:
*_MEG_Restin_preproc.zip
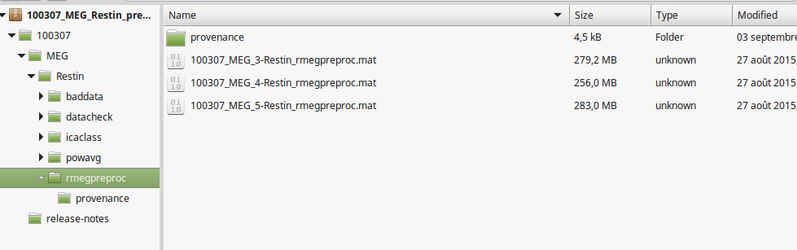
The folder structure can be explored by opening one zip file. Folder names are rather explicit (see right).
For instance, here, I want to retrieve all the resting state preprocessed MEG files. I can see that all of them contain the pattern *Restin_rmegpreproc.mat
rootdir = '/where/the/data/is'; destinationdir = '/where/the/data/goes'; % the patterns below should be adapted to capture specific zip files and specific files within it zippattern = '*_MEG_Restin_preproc.zip'; filepattern = '*-Restin_rmegpreproc.mat'; cd(rootdir) zips = dir(zippattern); cd(destinationdir) for i = 1:numel(zips) zipfname = fullfile(zips(i).folder,zips(i).name); system(['unzip -n ' zipfname ' ' filepattern]); end
In the zip file:
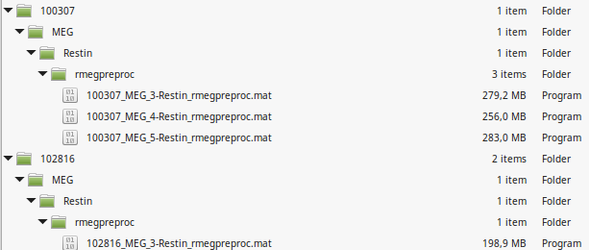
After unzip in your local folder:
Related articles
0 Comments