This is a collaborative space. In order to contribute, send an email to maximilien.chaumon@icm-institute.org
On any page, type the letter L on your keyboard to add a "Label" to the page, which will make search easier.
Organisational recommendations : subjects, scripts, where and how
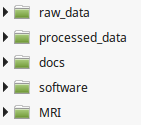
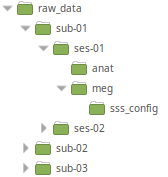
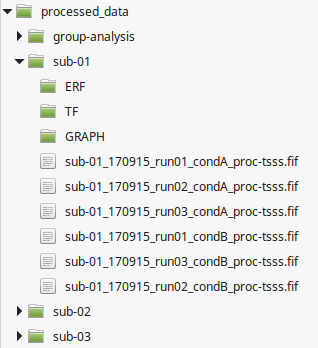
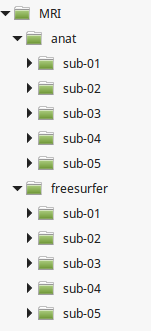
Here we describe the recommended organization for your data folder. It will ease the execution of the generic scripts you find in the wiki. It will also simplify the backup of your analysis. You are free to change this organization if your data doesn't fit in our generic framework.